by CJ Quines • on
The (real) systematic name of titin
it’s longer than everyone else thinks
If you’ve ever looked up “longest word”, you might’ve come across a word with 189,819 letters, starting with methionyl and ending with isoleucine. Or maybe you’ve watched the two-hour MrBeast video, where he claims to reads it. This word is, supposedly, the systematic name of titin, the largest known human protein.
Supposedly is the key word here. In fact, nearly all online sources that claim to have the name of titin are flat-out wrong, and the actual name is much longer, at 241,578 letters. Or maybe even longer, depending on what you consider to be titin!
How did I learn this? How do I know the other sources are wrong? And why does everyone else get this wrong?
Different sources
This year’s Galactic Puzzle Hunt had a puzzle about titin called Numbers.. My apologies for spoiling one aspect of the puzzle; if you want to give it a try, you should stop reading this post, before I spoil the rest of it.
This puzzle, which was by Lewis Chen, consists entirely of the following text:
Sources differ. Use Wiktionary to solve this puzzle.
9409
15414
49004
77205
82124
92209
96214
110120
126015
135520
136809
157620
161609
172514
The solution starts by looking at the last two digits of each number, and noticing that they’re all less than 26. Translating via A1Z26 spells INDEX INTO TITIN. The index here refers to indexing, a common puzzlehunt technique where you turn a number n into the nth letter of a word. Here, we index into the full name of titin, by taking the 9409th letter, the 15414th letter, the 49004th letter, and so on. This leads to a phrase, which leads to the answer.
Now, you might be curious about the sources differ note at the top. The authors’ notes in the solution tell the abbreviated story, which I’ll retell in more detail because I think it’s interesting. Here’s the timeline:
- June 7: Lewis posts the puzzle in our Discord server. Mark testsolves it and fails.
- June 8: Lewis updates the puzzle.
- July 2: Kevin Li testsolves the puzzle and succeeds.
- July 13: Brian Chen testsolves the puzzle and fails. On the last step, he gets
YYLLLRSCCYEAPY. - July 15: The puzzle gets solved during our Medium Big TestSolve.
- August 18: The puzzle and solution get postprodded, or put on the website.
- August 30: Brian starts factchecking the puzzle, making sure the puzzle and solution are correct. This is where it gets interesting.
That night, I think I was on a voice call with Brian, Mark, and some other people who were factchecking. Brian asked what source people used to get the name of titin, and Mark replied with a GitHub gist from hyperneutrino. Brian said he used a GitHub gist from drewchee, which differs from the first source after 36 letters.
For reasons that’ll become clearer later, let’s call the name in the first source name N2-B, and the name in the second source name F. Here’s where the difference starts:
methionylthreonylthreonylglutaminylalanylprolylthreonF
methionylthreonylthreonylglutaminylarginyltyrosylglut
Lewis claimed he used the Wiktionary name, which matches name N2-B. And different sources have different names. Brian compiled a spreadsheet of various sources:
| Source | Date | Name |
| Digital Spy | January 2024 | Name F |
| drewchee gist | July 2022 | Name F |
| hyperneutrino gist | April 2021 | Name N2-B |
| PDF on cw39 | October 2020 | Name F |
| Pokegym forum | May 2008 | Name F |
| Sarah McCulloch, 2009 | December 2009 | Name F |
| Sarah McCulloch, 2016 | December 2016 | Something else? |
| The Wild Life Blog | December 2021 | Name F |
| Wiktionary, 2010 | November 2010 | Name F |
| Wiktionary, 2011 | October 2011 | Name N2-B |
The name on Sarah McCulloch’s site is interesting—it starts like name N2-B, but it’s substantially longer. Given all this disagreement, we wanted to know: which name was correct? How would we even know?
The Blue Book
The source of truth for chemistry nomenclature is the International Union of Pure and Applied Chemistry, the IUPAC. There’s a series of books, the IUPAC Color Books, that are the authoritative references for these kinds of issues. The book for organic chemistry is the Blue Book, and the relevant section is P-103, amino acids and peptides.
Why is this relevant? We step into our first bit of chemistry: what is titin? Well, titin is a protein, which, for our purposes, is a chain of amino acids. This much might be familiar if you’ve taken a biology class. The unfamiliar word for me was peptide, which is also a chain of amino acids. Peptide is the general term, and proteins are particularly long peptides, usually at least fifty amino acids long. Peptides have an N-terminal and a C-terminal. By convention, peptide amino acid sequences are written from the N-terminal to the C-terminal.
Section P-103 doesn’t have the naming rules, but it instead refers to a separate document that does, the Nomenclature and Symbolism for Amino Acids and Peptides. Here we have the actual rules, 3AA-13, the naming of peptides:
To name peptides, the names of acyl groups ending in ‘yl’ (3AA-9.3) are used. Thus if the amino acids glycine, NH3+-CH2-COO-, and alanine, NH3+-CH(CH3)-COO-, condense so that glycine acylates alanine, the dipeptide formed, NH3+-CH2-CO-NH-CH(CH3)-COO-, is named glycylalanine. If they condense in the reverse order, the product, NH3+-CH(CH3)-CO-NH-CH2-COO-, is named alanylglycine. Higher peptides are named similarly, e.g. alanylleucyltryptophan. Thus the name of the peptide begins with the name of the acyl group representing the N-terminal residue, and this is followed in order by the names of the acyl groups representing the internal residues. Only the C-terminal residue is represented by the name of the amino acid, and this ends the name of the peptide.
This gives us the general structure of a systematic peptide name. The Wiktionary page refers to this in its etymology section. Both name N2-B and name F have this structure at the beginning:
methionylmethionine
threonylthreonine
threonylthreonine
glutaminylglutamine
alanylalanine
prolylproline
threonylthreonine
phenylalanylphenylalanine
methionylmethionine
threonylthreonine
threonylthreonine
glutaminylglutamine
arginylarginine
tyrosyltyrosine
glutamylglutamic acid
serylserine
Section 3AA-13 references 3AA-9.3, acyl groups, which references Blue Book Table 10.4, retained names of “common” α-amino acids, which I believe is the reference for the amino acid names. Name N2-B and name F first differ at the acyl groups corresponding to alanine and arginine.
During factchecking night, that August 30, Mark ruled out name F from being correct, as it’s not a valid protein name. The 269th acyl group is acetyl. This corresponds to acetic acid, which isn’t an amino acid. Name F also includes the substring titin, and it ends with serxisoluecine, both of which disqualify it from being a protein name. Hence my name: F for fake. I found this strange, given that name F was more widely circulated than name N2-B.
Was name N2-B valid, then? Manual inspection didn’t rule out the name, but to know for sure, we can write some code. First, the data for amino acid and acyl group names:
# Blue Book Table 10.4
amino_acids = { "A": "alanine", ... }# Blue Book Table 10.4
amino_acids = {
"A": "alanine",
"R": "arginine",
"N": "asparagine",
"D": "aspartic acid",
"C": "cysteine",
"Q": "glutamine",
"E": "glutamic acid",
"G": "glycine",
"H": "histidine",
"I": "isoleucine",
"L": "leucine",
"K": "lysine",
"M": "methionine",
"F": "phenylalanine",
"P": "proline",
"S": "serine",
"T": "threonine",
"W": "tryptophan",
"Y": "tyrosine",
"V": "valine",
}# 3AA-9.3
def to_acyl_group(amino_acid):
match amino_acid:
case "asparagine" | "cysteine" | "glutamine":
return amino_acid.removesuffix("e") + "yl"
case _ if amino_acid.endswith("ic acid"):
return amino_acid.removesuffix("ic acid") + "yl"
case _ if amino_acid.endswith("an"):
return amino_acid.removesuffix("an") + "yl"
case _ if amino_acid.endswith("ine"):
return amino_acid.removesuffix("ine") + "yl"
case _:
raise Exception
acyl_groups = {
symbol: to_acyl_group(amino_acid)
for symbol, amino_acid in amino_acids.items()
}Then, parsing a protein name. Protein names can only be a sequence of acyl group names followed by an amino acid name; there’s no alternatives or anything. This makes the parser simpler than a square dance parser I previously wrote about.
I had to be a bit more efficient than that parser though, especially since we’re parsing a long string. A simple approach would be to take out the acyl group name from the start of the protein name and then parse the rest. This can be accidentally quadratic, however, if you copy the string each time. It’s faster to only keep track of indices:
def parse_acyl_group(word, i):
for symbol, acyl in acyl_groups.items():
if word[i : i + len(acyl)] == acyl:
return symbol
def parse_amino_acid(word, i):
for symbol, acid in amino_acids.items():
if word[i : i + len(acid)] == acid:
return symbol
# 3AA-13
def protein_to_amino_acids(word):
i = 0
while i < len(word):
if symbol := parse_acyl_group(word, i):
yield symbol
i += len(acyl_groups[symbol])
elif symbol := parse_amino_acid(word, i):
yield symbol
i += len(amino_acids[symbol])
else:
break
assert i == len(word)With this, we can check that name F is indeed an invalid protein name; it fails the assertion at the end. We can check that name N2-B is a valid protein name.
Sequence conflict
To recap: name N2-B is a valid protein name, and we’ve ruled out name F. These were, by far, the two most common titin names floating around the internet, so we can be reasonably confident that name N2-B is correct, right? Unsatisfied with the uncertainty, and with a puzzle that still needed to be factchecked, Brian came back to settle some questions.
On September 13, Brian tried to generate titin’s name himself. The UniProt entry for titin, Q8WZ42, contained several sequences. Apparently proteins can have isoforms, which are different variants of the same protein. Let me explain real quick.
The gene for a protein has several exons, which are segments of the gene that, when spliced together and translated, form the protein. By omitting some of the exons, we get different isoforms of the same protein; this is called alternative splicing. Alternative splicing is only one way that protein isoforms are made, and it’s the one that produces different titin isoforms.
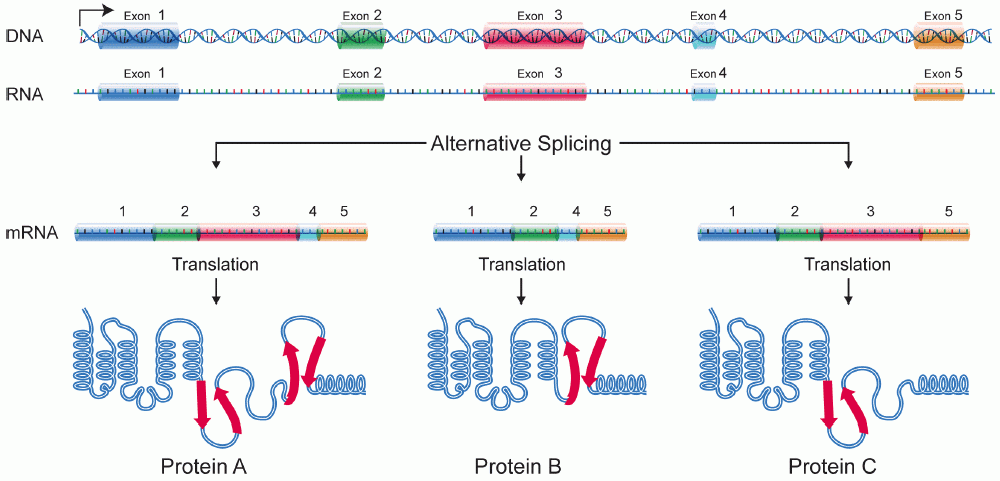
The UniProt entry lists thirteen different isoforms, one which was chosen as canonical. You can download its amino acid sequence; it’s a long text file of amino acid symbols, starting with MTTQAPTFTQ. We can use our same amino acid name data to translate this to a name:
def amino_acids_to_protein(symbols):
*head, tail = symbols
name = []
for symbol in head:
name.append(acyl_groups[symbol])
name.append(amino_acids[tail])
return "".join(name)Brian did this, and got a name that was 241,578 letters long. Let’s call it name N2-BA. The next day, I independently reproduced his work—resulting in the code you see above—and got the exact same result.
I was in utter disbelief. How were people not only wrong with their so-called longest word, but short by more than fifty thousand letters? Well, it turns out that name N2-BA appears on the internet, though on only one source: Sarah McCulloch’s website. More on external sources later; for now, let’s focus on the names.
If name N2-B wasn’t the name of the canonical isoform of titin, then what was it a name of? Brian discovered that the sequence for name N2-B was quite similar to one of the isoforms listed on UniProt, small cardiac N2-B, hence the name. Its amino acid sequence was nearly the same as name N2-B’s, but they differed in 59 amino acids. And to be clear, that’s only 59 amino acids, out of 26,926. That was too similar to be coincidence.
Turns out that UniProt has a peptide search. I searched up the protein corresponding to name N2-B, and got an exact match on UniParc, UPI000011010D. Its UniProtKB entry was a greyed out Q10466, and it’s greyed out because it was merged with the entry Q8WZ42. Which was the UniProtKB entry for titin I linked earlier. Wait, what?
In the UniProtKB entry for titin, under a subsection called “Sequence conflict”, the sequence for name N2-B is listed under the ID CAA62188.1. The listed reason for their difference is a frameshift. That settled the matter then. Name N2-B is the systematic name for a frameshifted version of N2-B, which is an isoform of titin, but not the canonical one. Whether or not name N2-B is a “correct” name depends on whether you count frameshifts or non-canonical isoforms.
The frameshift, and other possible titin mutations, could be somewhat common, so perhaps it’s not disqualifying. In the /r/labrats subreddit, under a “titin hate thread”, /u/dusting_for_vomit commented:
I work in clinical genetics and I hate TTN too! (though for different reasons). Due to its sheer size, we encounter untold numbers of missense variants in TTN during diagnostic testing that are rare or completely novel in the general population. Even though such variants are unlikely to be disease associated, we cannot rule out a potential disease risk simply because we have never seen them before, and thus they get reported back to patients and clinicians as “variants of uncertain significant”. This is annoying because it makes clinical reports more complicated than they need to be. Screw that gene.
Even if you think mutations disqualify name N2-B from being correct, it’s not clear that name N2-BA is correct. In particular, UniProt’s canonical titin sequence might not be so canonical. The other candidate canonical isoform is listed in UniProt under a different entry, C9JQJ2. While UniProt doesn’t have much to say about it, its RefSeq entry NP_001254479.2 calls it the IC isoform. Name N2-BA is in RefSeq under NP_001243779.1, where it’s called the N2-BA isoform, hence the name.
The special thing about IC is noted in its RefSeq entry: it’s an inferred transcript of titin. N2-BA is directly attested in the human body, per article PMID 10850961. But IC is a predicted protein, formed by putting together all the exons in the titin gene; as far as I can tell, it’s not a protein that’s been found in humans. While N2-BA is 34,350 amino acids long, IC is 35,991 amino acids long. I found a page about titin exons, because of course there’s a whole page about titin exons, and it shows which exons in IC are also present in the other isoforms.
So, is IC a more “canonical” version of titin than N2-BA? It’s certainly longer; the systematic name of IC, name IC, has 252,178 letters. But why stop there? We know of longer proteins in other species, and possibly more we haven’t discovered. In any case, if you want the systematic name for the largest protein that appears in the human body, you probably want the 241,578-letter name N2-BA.
Verifiability
It’d be a much cooler story if no one had noticed this before, but that’s not the case. I can find name N2-BA referenced in at least four places on the internet:
-
The most recent is in the solution to the puzzle that spawned all this in the first place.
-
I’ve already mentioned Sarah McCulloch’s site, which, at the time of writing, gives the correct name N2-BA but cites the incorrect length. This was because it originally had name F, and the post was corrected after a comment from Stephen Thomas in December 2016. Stephen, under the username flabdablet, also references name N2-BA in a forum post on The Daily WTF forums.
The comment, and the discussion on the forum thread, mention that the isoform Q8WZ42-12 is larger than N2-BA. This isoform corresponds to RefSeq entry NP_00125479.1. This is version 1 of the entry for IC; the latest version, version 2, differs in one amino acid. Though, as I’ve mentioned earlier, I don’t believe that IC is a protein found in the human body.
-
There’s also a blog post from Alex Howe, made in September 2022, that covers a lot of what this blog post does as well.
-
I’ve found that the most interesting references to name N2-BA, however, are in the Wikipedia talk page for titin. Let’s take a look…
A lot of the discussion sections center on whether to include the name of titin on the page. There’s some discussion from September 2010 about different isoforms, and a topic from October 2016 mentions the difference between name N2-B and name F.
The interesting discussion topic is “Linguistic significance”. I can’t tell exactly what happened in the discussion here; there’s a reply dated May 2015 to a comment dated June 2017, so there must be some editing that got messed up. Anyway, the comments from IvanP and Stuart M Klimek, together, cover a lot of the things I’ve mentioned in this post. Both commenters say that the article has incorrect information. This incorrect info persists even in the current version of the Wikipedia page:
As the largest known protein, titin also has the longest IUPAC name of a protein. The full chemical name of the human canonical form of titin, which starts methionyl… and ends …isoleucine, contains 189,819 letters and is sometimes stated to be the longest word in the English language, or of any language.
As we now know, titin is not the largest known protein, there’s several possible canonical isoforms of titin, and all of them have names longer than 189,819 letters. And yet, the Wikipedia page still has this incorrect information, even after the discussion in the talk page. Why?
The answer has to do with Wikipedia’s content policies, particularly WP:V, verifiability. Information in Wikipedia articles doesn’t have to be true. Rather, it needs to be verifiable, which means that people can find this information on a published reliable source. This means that if something false has been published in several reliable sources, then it’s fine; it can go on Wikipedia.
What counts as a reliable source? In general, there’s a spectrum of reliability, and the requirement for how strong sources need to be becomes higher when the information is likely to be challenged. The cited source for the claim that titin’s name is 189,819 letters is McCulloch’s site. While it’s a self-published source, the same information is repeated by stronger sources, like Digital Spy, Christian Science Monitor, HuffPost, and PBS, all of which are considered perennial sources.
In contrast, the claims that name F is wrong, or that name N2-B is only the name of an isoform of titin, or that name N2-BA is longer and has so-and-so many letters, are all claims that only appear in self-published sources. As we’ve seen above, these claims have appeared in blog posts, forum posts, and comments on Wikipedia talk pages, which are all excluded per the policy WP:NOR, no original research. Maybe these sources would count if they were produced by subject-matter experts, but I don’t plan on becoming a bioinformatician any time soon.
What if I wanted to get the fact removed from Wikipedia? Or replaced with correct information? Then I’d have to work to get stuff published in more reliable sources than the incorrect information. And for a fact as unimportant as this, why does it matter if the Wikipedia page gets it wrong, or if other Wikipedia pages get it wrong too?
Citogenesis
In 2011, Randall Munroe coined citogenesis in an xkcd comic. The term refers to circular reporting, where incorrect information on Wikipedia is repeated by a reliable source, and that source is then used as a citation for the incorrect information.
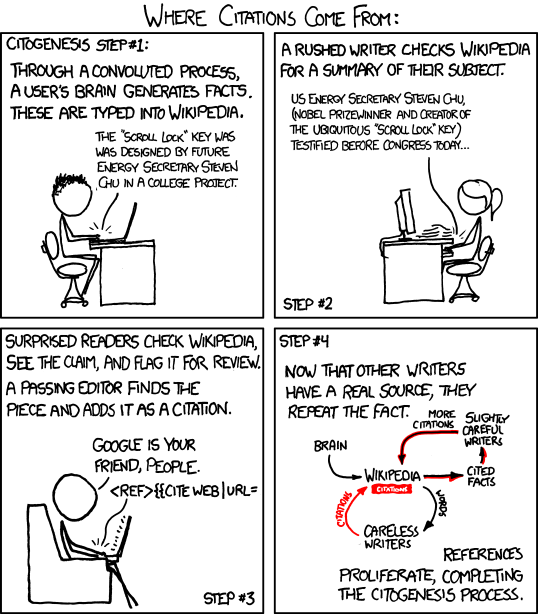
I think that the propagation of name N2-B, and later name F, is an example of citogenesis. Here’s a timeline:
1970s. The Guinness Book of World Records, 18th edition, cites the systematic name of tryptophan synthesase as the longest chemical word. The history of that name is covered elsewhere; see a different blog post from Alex Howe.
March 2005. Wikipedia user Brian0918 generated the systematic name for the protein enaptin, using the amino acid sequence on UniProt, and posts it to Wikisource. He then made the Wikipedia article for enaptin and edited the longest word in English article, which was, presumably, what inspired Brian0918 to do all this.
A few days later, Slashdot user timothy posts a story about the enaptin article. A comment from user syntax asks about the longer protein titin. Brian0918 generates name N2-B, posts it on Wikisource, creates the the Wikipedia article for titin, and edits the longest word in English article.
As an aside, the two-hour Mr. Beast video that claims to read the name of titin actually reads the name for enaptin. The systematic name for enaptin is 64,060 letters, which is less than a third of the length of name N2-B. Maybe he should reshoot the video with the correct name?
June 2006. The Wikisource pages with the systematic names of enaptin and titin are proposed for deletion. When the latter page was deleted, it still had name N2-B.
September 2006. Wikipedia user Convoy of Conwy edits the Wikipedia titin article to add name F. This is the earliest appearance I could find for name F, and I wish I knew how Convoy of Conwy got this name.
Stephen Thomas commented on McCulloch’s post with a shell script that generates name F, showing that it’s quite repetitive. So one possible explanation is that Convoy of Conwy (or whoever originally posted name F) made a huge error while copying and pasting the name around.
A few months after first publishing this post, Ivan Panchenko wrote in with the observation that Name F is similar to two names on one of Jeff Miller’s long words pages. In particular, we can rewrite Stephen Thomas’s program in terms of these names:
tmv = "acetyl[…]serine" # tobacco mosaic virus
tsa = "methionyl[…]serine" # tryptophan synthesase A
A = "ethionylthreonylthreonyl" + tsa[9:1106]
B = tsa[1106:1478]
C = tsa[1478:-6] + tmv[:771]
D = tmv[771:-6] + "titin" + tsa[:70]
E = tmv[727:769] + "yl" + tmv[727:769] + "xisoleucine"
F = B + C + D
G = A + 4 * F
H = "m" + G
print(10 * H + G + 9 * H + C + D + C + E)August 2007. After several months of name N2-B and name F getting removed and added from the Wikipedia page, Othyr posts name F on their website. They comment on the talk page, claiming to have gotten the name from “what had been deleted in the article”. This link is added to the article as an external link.
From here, name F begins to propagate. At this point, the only source for name N2-B is in the history of the Wikipedia article, but name F is on Othyr’s site, which is linked in the article. Name F then gets copied to several more places.
November 2010. Wiktionary user Liliana-60 creates the Wiktionary page with name F.
October 2011. Wiktionary user -sche, while adding details like pronunciation and etymology to the Wiktionary entry, somehow discovers that name F is wrong, and adds name N2-B. This is the next appearance I could find of name N2-B on the internet; before that, it was only in the Wikipedia article history.
Anyway! This is only one of many citogenesis incidents. Perhaps less consequential than some other incidents, though it did lead to someone spending three-and-a-half hours reading name F. Which isn’t even a correct protein name.
Thanks to Brian for editing.